The first principal component.
Illustration from Pearson's paper.
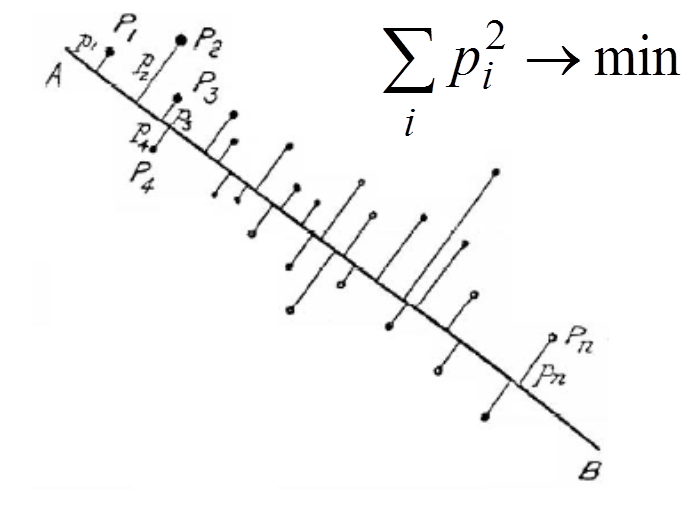
Data points are xi (i=1,...,m). In the applet, they are situated on the 607ū400 workdesk.
|
The first principal component. |
Two initial nodes, y1, y2, are defined by the user before learning. The data points xi (i=1,...,m) are given.
The Self-Organizing Map (SOM), also known as the Kohonen network, is a computational method for the visualization, low-dimensional approximation and analysis of high-dimensional data. It works with two spaces: a low-dimensional space with a regular grid of nodes and the higher-dimensional space of data. In our applet, the data space is two-dimensional (a plane) and the nodes form a one-dimensional array.

Radius 3 linear B-spline |
Each node ni is mapped into the dataspace. Its image, yi is called the coding vector, the weight vector or the model vector. Rather often, the images of nodes in the data space are also called "nodes", with some abuse of language, and the same notation is often used for the nodes and the corresponding coding vectors. The exact sense is usually clear from the context.
The data point belongs to (or is associated with) the nearest coding vector. For each i we have to select data points owned by yi. Let the set of indices of these data points be Ci: Ci={l:||xl-yi||2≤||xl-yj||2 ∀i≠j} and |Ci| be the number of points in Ci.
The learning of SOM is a special procedure that aims to improve the approximation of data by the coding vectors and, at the same time, approximately preserves the neighbourhood relations between nodes. If nodes are neighbours then the corresponding coding vectors should also be relatively close to each other. To improve the approximation of data, each coding vector yi moves during learning towards the mean point of the data points owned by yi. It involves in this movement the neighbours to preserve neighbourhood relations between nodes. This Batch SOM learning rule was proposed by Kohonen. The learning algorithm has one parameter, the "Neighbourhood radius" hmax. It is necessary to evaluate the neighbourhood function and this should be assigned before learning. For this applet, the neighbourhood function, hij, has the simple B-spline form: 1-hij=|i-j|/(hmax+1) if |i-j|=hmax and hij=0 if |i-j|>hmax (the example is given on the right).
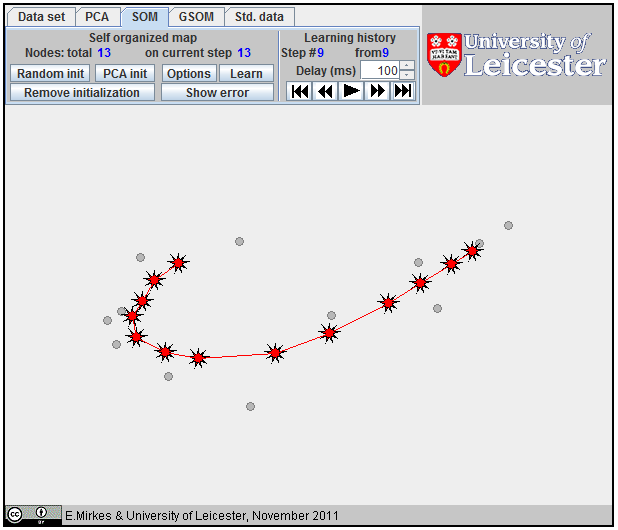
1D SOM as a broken line. |
The initial location of coding vectors should be assigned before the learning starts. There are three options for SOM initializations:
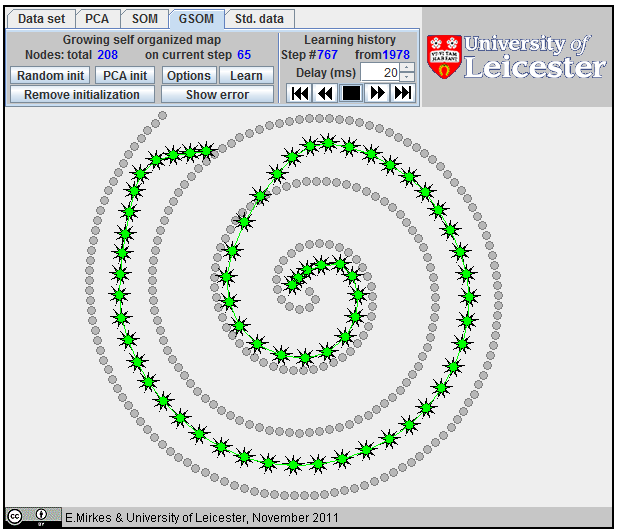
Approximation of a spiral by GSOM; 65 nodes, 
Approximation of a spiral by GSOM; 152 nodes. 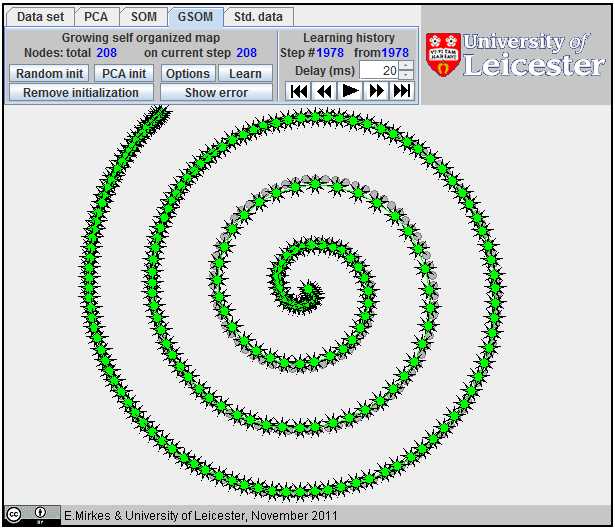
Approximation of a spiral by GSOM. 
All attempts to approximate a spiral by |
The initial location of coding vectors should be assigned before the learning starts. There are three options for GSOM initializations:
All learning steps (vectors yi) are saved into "Learning history". If learning restarts then yi from the last record of the Learning history are used instead of user defined initial coding vectors.
The applet contains several menu panels (top left), the University of Leicester label (top right), work desk (center) and the label of the Attribution 3.0 Creative Commons publication license (bottom).
To wisit the web-site of the University of Leicester click on the University of Leicester logo.
To read the Creative Commons publication license click on the bottom license panel.
|
Common parts of the menu panel for all methods
The All methods menu panel containes two parts.

|
|

|
For PCA, all points and lines are in a blue color. The points for PCA have the form of four-ray stars (see left). To start PCA learning two points are needed. You can select initial points by mouse clicks at choosen locations, or by the button Random initialization. When you try add the third point, the first point is removed.
After learning is finished, use the Learning history buttons at the right part of menu panel to look at the learning history and results. You will see one four-ray star, the centroid of the data set, and one line, the current approximation of the first principal component.

|
For SOM, all points and lines are in a red color. The points for SOM have the form of nine-ray stars (see left). To start SOM learning, the initial positions of all coding vectors are needed. To add a point to an initial approximation, click the mouse button at the choosen location, or use the button Random init or the button PCA init. When you add a new point, it is attached to the last added point.
After learning is finished, use the Learning history buttons at the right part of menu panel to look at the learning history and results.

|
For GSOM, all points and lines are in a green color. The points for GSOM have the form of nine-ray stars (see left). To start learning, GSOM is needed to initiate one point. To place initial point click the mouse button in the chosen location, or push the button Random init or the button PCA init. When you add a new point, the new point is joined with the last added point.
After learning is finished, use the Learning history buttons at the right part of menu panel to look at the learning history and results.
|
The distance from a point to a straight line is the length of a perpendicular dropped from the point to the line. This definition allows us to evaluate FVU for PCA. For SOM and GSOM we need to solve the following problem. For the given array of coding vectors {yi} (i=1,2, ... m) we have to calculate the distance from each data point x to the broken line specified by a sequence of points {y1, y2, ... ym}. For the data point x, its projection onto the broken line is defined, that is, the closest point. The square of distance between the coding vector yi and the point x is di(x)=||x-yi||2 (i=1,2, ... m).
|
Projection of a data point onto a segment |
Let us calculate the squared distance from the data point x to the segment [yi, yi+1] (i=1,2, ... m-1). For each i, we calculate li(x)=(x-yi , yi+1-yi ) /||yi+1-yi||2 (see left).
If 0<li(x)<1 then the point, nearest to x on the segment [yi, yi+1], is the internal point of the segment. Otherwise, this nearest point is one of the segment's ends.
Let 0<li(x)<1 and c be a projection of x onto segment [yi, yi+1]. Then ||c - yi||2=(li(x)||yi-yi+1||)2 and, due to Pythagorean theorem, the squared distance from x to the segment [yi, yi+1] is ri(x)=||x-yi||2-(li(x) ||yi-yi+1||)2.
Let d(x)=min{di(x) | i=1,2, ... m} and r(x)=min{ri(x) | 0< li(x) <1, 0< i< m }. Then the squared distance from x to the broken line specified by the sequence of points {y1, y2, ... ym} is D(x)=min{d(x), r(x)}.
For the given set of the data points xj and the given approximation by a broken line, the sum of the squared distances from the data points to the broken line is S=∑jD(xj), and the fraction of variance unexplained is S/V, where V=∑j(xj - X )2 and X is the empirical mean: X=(1/n)∑xi.